FAU researchers shine a spotlight on an important risk factor for Parkinson’s and Gaucher’s disease
In Germany, approximately 400,000 people suffer from Parkinson’s, and numbers are on the rise. Using an electron microscope, a team of researchers at Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU) and Uniklinikum Erlangen (UKER) were able to make a major risk factor for the diseases visible: the structure of the GCase enzyme in conjunction with its transport protein LIMP-2, which transports the enzyme through the cell to its site of action.
Mutations in the enzyme β-glucocerebrosidase (GCase) increase the probability of developing Parkinson’s disease by approximately 20 times. These mutations are also the cause of the rare metabolic disorder Gaucher’s disease. Together with partners from Osnabrück, Belgium and the USA, the FAU/UKER research team led by Prof. Dr. Friederike Zunke and PD Dr. Philipp Arnold has succeeded in making the structure of the GCase enzyme visible together with its transport protein LIMP-2. They used a cryogenic electron microscope that makes the protein structure visible at an atomic resolution.
Protein structure as a risk factor for Parkinson’s and Gaucher’s disease
It is a major problem if a person is lacking the GCase enzyme or if the enzyme does not function correctly. “An enzyme is a protein with a particular task in the body,” explains Prof. Friederike Zunke. “In the case of GCase, this enzyme is responsible for breaking down a certain substrate that is a lipid, or, putting it simply, a fat.” If the enzyme fails to break down the substrate this accumulates in the cells together with other protein aggregates. This is particularly problematic when it affects cells that do not split. “If a cell splits, then its content also splits,” explains Friederike Zunke. “If I have a protein aggregate, a clump of proteins, in the cell, then this would split together with the cell and the problem would probably be less serious. However, our nerve cells no longer split, and that is why protein aggregates collect there if the enzyme fails to work.”
This happens, for instance, with Gaucher’s disease, a lysosomal storage disease. Lysosomes are sac-like cell organelles encompassed by a membrane. Since they contain various enzymes that they use to break down foreign substances or bodily waste products, they are also known as the body’s recycling depot. In Gaucher’s disease, the lysosomes store the GCase substrate that cannot be broken down, clogging the cells as a result. This leads to an accumulation of waste products in the cells, which leads to a gradual dying off.
Furthermore, GCase is one of the greatest genetic risk factors for Parkinson’s disease. If GCase has certain mutations, it can increase the probability of developing Parkinson’s disease by up to twenty times. “This explains why maintaining the enzyme activity of GCase is an important component when researching new treatment options for Parkinson’s disease,” explains Friederike Zunke. Studies have shown that fewer damaging aggregates accumulate if the GCase enzyme is activated. “This principle was tested for various activators in clinical studies. The results underline the importance of a detailed understanding of the structure of this protein and its transporter,” explains Philipp Arnold.
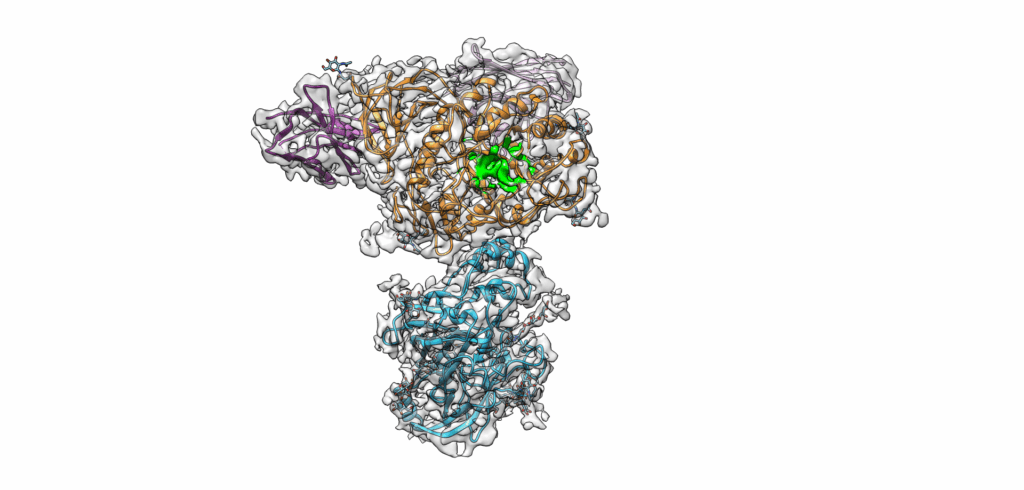
New research approaches: Can the enzyme be activated naturally?
The research team led by Friederike Zunke and Philipp Arnold not only succeeded in making the molecule visible but also clarified how the enzyme GCase and the transport protein LIMP-2 interact. “We observed that the enzyme is only activated by binding to its transporter LIMP-2. In future, now that we have decoded the structure of the transport complex, this may lead to options for developing new treatments that attach to exactly this location on the protein,” explains Friederike Zunke. However, a lot of research remains to be done.
Original publicationCryogenic electron microscopy
In cyrogenic electron microscopy, a protein solution is frozen so quickly in its natural environment that no ice crystals can form. The proteins are essentially captured in the glass-like ice in their natural surroundings. Using the electron microscope, images are taken at high resolutions (in our case at 130,000 times), and as the proteins were able to move freely until shortly before they were flash-frozen, it is possible to take images of them from all orientations. Approximately 10,000 images are taken per dataset, and then suitable algorithms are used to cut out individual protein particles and save them in a gallery. Such image galleries can include several million particles. In the next step, particles showing the same view of the protein are sorted and summed up in one group. This amplifies the protein signal and reduces the unspecific background. All class sums obtained in this way are now assigned angles in order to be able to calibrate them to each other in a 3-dimensional space. Projecting the information from the two class sums along their axes, reveals the structure of the protein where the projections meet. The method of calculating the structure on the basis of many individual protein particles is called single particle analysis and was awarded the Nobel Prize in 2017. Nowadays, resolutions of under 2Å (0.2 nm) can be obtained under the correct circumstances.
Further information:
Prof. Dr. Friederike Zunke
Professorship for Translational Neurosciences
Department of Molecular Neurology, UKER
Friederike.zunke@fau.de
PD Dr. Philipp Arnold
Institute of Functional and Clinical Anatomy
Philipp.arnold@fau.de

